Real-time 2D skeletonization using CUDA
2D skeletonization is becoming an essential tool in the image processing practitioner's toolbox. Methods exist which guarantee robustness, accuracy, resistance to noise, correctness, and completeness properties. However, performance is still a major issue, especially when processing huge datasets such as videos or image collections.
Real-time CUDA skeletonization
We present here a fast adaptation of the augmented fast marching method (AFMM) for computing 2D skeletons. For this, we implement the AFMM skeleton detector and simplification principle, which ensures correctness, noise resistance, and completeness, within a GPU setting using CUDA. To our knowledge, to date, this is the world's fastest continuous-multiscale 2D skeletonization method. Our method works as follows.
Distance and feature transforms
Given a binary pixel shape, we compute the distance transform (DT) and one-point feature transform (FT). For this, we use an optimized version of the parallel banding algorithm. Storing the FT delivers the exact Euclidean distance to boundary, which we can next compute using floating-point or other number representation schemes, as desired. Thresholding the DT delivers exact shape dilations and erosions.


The image above-left is the DT for a leaf shape (contour in red), encoded in grayscale. The image above-right shows the FT. Boundary pixels are labeled monotonically from black to white. Point colors in the FT indicate the closest boundary pixel (for details on this scheme, see here).
Skeleton importance
With the FT, we compute the skeleton importance using the proven criterion introduced by the AFMM. This operation is implemented as a single, efficient, image pass in CUDA.
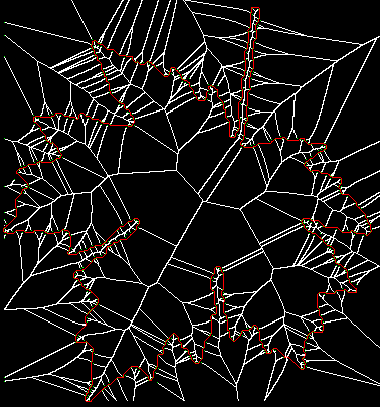

The images above show the full skeleton of the leaf (left) and a simplified version thereof (right). Skeletons for both inside and outside the shape are computed. If desired, one of these can be eliminated by simple masking.
Skeleton endpoint detection
Many imaging applications require finding the endpoints of skeleton branches e.g. for shape matching or analysis. We implement this in CUDA using a 3 by 3 template in a single image pass. The method delivers both a labeling of the skeleton endpoints and an explicit endpoint array. The 'gather' step is done efficiently using CUDA 1.1's atomicInc operation.
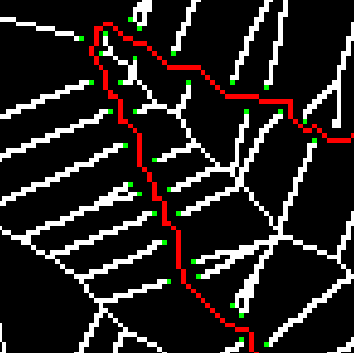
The image above shows a zoom-in of the leaf skeleton. Green pixels indicate endpoints.
Area filling
Prior to skeletonization, noisy images may need hole filling to remove small-scale artifacts. Doing this by dilation and erosion in our toolkit is possible, but suboptimal performance-wise when holes are large. We propose a new general-purpose scan-line method for area filling in digital images. The method uses an interleaved x-y scan-line approach which parallelizes efficiently on CUDA. For details, see the source code below.
Cushion shading
Skeletons and DTs can be used to generate cushion shading profiles for any closed 2D shapes. For this, we interpolate between the DT of the shape and the DT of the skeleton, and regard the result as intensity. The image below presents a cushion shading of the leaf: pixels close to the leaf center (skeleton) are bright, while pixels close to the boundary are dark. This technique was used to generate simplified renderings of large bundled graphs.

Performance
Below are timings for an image of 512x512 pixels on a MacBook Pro with an Nvidia GT 330M card:
- DT and FT: 4.0 milliseconds
- skeleton: 0.27 milliseconds
- endpoints: 0.5 milliseconds
- hole filling: 3.1 milliseconds
Overall, our method is 20..80 times faster than the original CPU-based AFMM. The method is linear in the number of pixels. The attained performance makes it applicable to real-time video processing.
Implementation
Our skeletonization toolkit is implemented as a C++ library using CUDA 1.1. The source code is available for research purposes here. To use it
- build the software (a makefile for Linux/Mac OS X is provided)
- run the demo application
skeletonfilename.pgm. Inputs are binary images (black=inside, white=outside), see the DATA directory. - read the README file for navigation options
- modify the main file to integrate the code in other applications, as needed
Applications
We used our CUDA skeletonization toolkit in several applications, among which we note simplified bundled graph visualization and general graph bundling.
