Skin Lesion Segmentation with Dense Skeletons
The diagnosis and prognosis of skin tumors, such as naevi or melanoma, is an increasingly important problem. Given the growing prevalence of such tumors, designing efficient and effective methods to analyze imaged skin lesions is an important goal, e.g. for the automatic measurement of the ABCD diagnostic score.
Tumor Segmentation: Requirements
A key part of tumor image analysis is the segmentation of the tumor from the surrounding regular skin tissue. This is however a hard, since a tumor may
- have weak-contrast, fuzzy, complex-shaped borders
- consist of several types of sub-patterns (e.g. network, globular, unstructured, or salt-and-pepper)
- be partially overlapped by hair structures
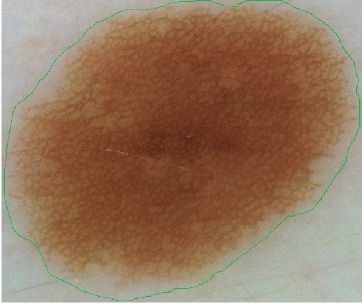
The image above shows a reticulated-pattern tumor. The green contour indicates a manual segmentation performed by a dermatologist. Note how the contour
- surrounds the brown area at a certain distance
- has a smooth appearance, ''i.e.' does not follow all tumor details
Tumor Segmentation by Dense Skeletons
We developed a method to segment such skin lesions based on a new image representation: dense skeletons. The outline is:
- extract all luminance-based, or hue-based, structures from an image (using threshold sets)
- simplify each structure using saliency skeletons
- measure the relevance of each simplified structure to the entire image
- segment the image by choosing the most relevant structures
The images below show two such possible segmentations of the tumor in the image above:
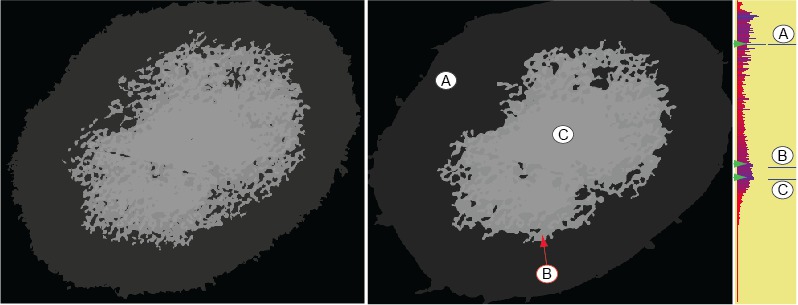
Left image: The tumor has been segmented here into two structures. The outer structure (light gray) corresponds quite well with the outline manually drawn by the dermatologist. The inner structure (dark gray) corresponds to the core of the tumor, i.e. the darker-brown reticulated structure in the middle.
Middle image: The tumor has been selected here into three structures (A, B, C). The right bar-plot shows the relevance values for all 255 threshold-sets in the image. Note how the levels corresponding to the three salient structures A, B and C correspond to local relevance maxima. This allows detecting, and thus segmenting, such images using a simple automatic local maxima detection.
Experimental validation
To validate our method, we compared our automatic segmentation results with 50 images acquired from different types of melanoctyic lesions. The images cover a variety of lesion patterns (network, globular, unstructured, and salt-and-pepper) and hues, and have varying degrees of hair occlusion. Images were acquired with a Handyscope at the resolution of 2448 x 3264 pixels. Images were next segmented manually by a dermatologist, using a simple paint program to delineate the tumor. For automatic segmentation, we selected the largest relevant segment, corresponding to the tumor outline. The two segmentations were done double-blind. Visual comparison of the segmentation results showed a very good match of the manual and automatic segmentation.
Implementation
Our segmentation method is implemented atop of the dense-skeleton image representation using a CUDA-based salience skeleton implementation. The method runs at approximately 1 image/second at the Handyscope full resolution on a MacBook Pro with a Nvidia 330 GTM graphics card.
Publications
Several publications related to this project are listed below.
- Automatic segmentation and visualization of skin lesions (R. Kiers, J. Jans, A. Telea, research report PDF)
- A Dense Medial Descriptor for Image Analysis (M. van der Zwan, Y. Meiburg, A. Telea, Proc. VISAPP, 2013)
- Automatic Segmentation of Skin Lesions Using Multiscale Skeletons (D. Boda, A. Diaconeasa, S. Zurac, A. Telea, M. Neagu, C. Constantin, C. Solovan, R. Voinescu, Proc. 8th EADO Congress, 2012)
- Qualitative Comparison of Multiscale Skin Tumor Segmentation Methods (D. Boda, A. Diaconeasa, M. Neagu, C. Solovan, A. Telea, R. Voinescu, S. Zurac, Proc. 4th Intl. Conf. of the Romanian Society of Dermato-Oncology, 27-29 June 2013, Cluj, Romania)
- Color Tunneling: Interactive Exploration and Selection in Volumetric Datasets (C. Hurter, A. R. Taylor, S. Carpendale, A. Telea, Proc. IEEE PacificVis 2014)
Partners and acknowledgments
This research was done in collaboration with Daniel Boda MD PhD and Adriana Diaconeasa MD PhD, Center of Excellence in Dermato-oncology, Univ. of Medicine and Pharmacy Carol Davila, Bucharest, Romania. The work is financed by the research grant PN-II-RU-TE-2011-3-0249 of the Ministry of Education, Research, and Sports (MECTS/UEFISCDI), Romania.
